Cephalosporin resistance, tolerance, and approaches to improve their activities | The Journal of Antibiotics - Nature.com
Abstract
Cephalosporins comprise a β-lactam antibiotic class whose first members were discovered in 1945 from the fungus Cephalosporium acremonium. Their clinical use for Gram-negative bacterial infections is widespread due to their ability to traverse outer membranes through porins to gain access to the periplasm and disrupt peptidoglycan synthesis. More recent members of the cephalosporin class are administered as last resort treatments for complicated urinary tract infections, MRSA, and other multi-drug resistant pathogens, such as Neisseria gonorrhoeae. Unfortunately, there has been a global increase in cephalosporin-resistant strains, heteroresistance to this drug class has been a topic of increasing concern, and tolerance and persistence are recognized as potential causes of cephalosporin treatment failure. In this review, we summarize the cephalosporin antibiotic class from discovery to their mechanisms of action, and discuss the causes of cephalosporin treatment failure, which include resistance, tolerance, and phenomena when those qualities are exhibited by only small subpopulations of bacterial cultures (heteroresistance and persistence). Further, we discuss how recent efforts with cephalosporin conjugates and combination treatments aim to reinvigorate this antibiotic class.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
References
Marshall WF, Blair JE. The cephalosporins. Mayo Clin Proc. 1999;74:187–95.
Brotzu G. Research on a new antibiotic. Cagliari Inst Hyg. 1948;1:5–15.
Newton GG, Abraham EP. Cephalosporin C, a new antibiotic containing sulphur and D-alpha-aminoadipic acid. Nature. 1955;175:548.
Abraham EP. Cephalosporins 1945–1986. Drugs. 1987;34:1–14.
Chauvette RR, et al. Chemistry of Cephalosporin Antibiotics .2. Preparation of a new class of antibiotics and relation of structure to activity. J Am Chem Soc. 1962;84:3401.
Godzeski CW, Brier G, Pavey DE. Cephalothin, a new cephalosporin with a broad antibacterial spectrum. I. In vitro studies employing the gradient plate technique. Appl Microbiol. 1963;11:122–7.
Griffith RS, Black HR. Cephalothin-a new antibiotic. Preliminary clinical and laboratory studies. JAMA. 1964;189:823–8.
Chang TW, Weinstein L. In vitro biological activity of cephalothin. J Bacteriol. 1963;85:1022–7.
WHO model list of essential medicines - 22nd list. World Health Organization, Document: WHO/MHP/HPS/EML/2021.02 (2021).
The WHO AWaRe (Access, Watch, Reserve) antibiotic book. World Health Organization, Document: ISBN 978-92-4-006238-2 (2022).
Goldstein E. Rise in the prevalence of resistance to extended-spectrum cephalosporins in the USA, nursing homes and antibiotic prescribing in outpatient and inpatient settings. J Antimicrob Chemother. 2021;76:2745–7.
Penalva G, et al. Decreasing and stabilising trends of antimicrobial consumption and resistance in Escherichia coli and Klebsiella pneumoniae in segmented regression analysis, European Union/European Economic Area, 2001 to 2018. Euro Surveill. 2019;24:1900656.
Choby JE, Ozturk T, Satola SW, Jacob JT, Weiss DS. Does cefiderocol heteroresistance explain the discrepancy between the APEKS-NP and CREDIBLE-CR clinical trial results? Comment. Lancet Microbe. 2021;2:E648–E649.
Choby JE, Ozturk T, Satola SW, Jacob JT, Weiss DS. Widespread cefiderocol heteroresistance in carbapenem-resistant Gram-negative pathogens. Lancet Infect Dis. 2021;21:597–8.
Jia X, et al. Heteroresistance to cefepime in Pseudomonas aeruginosa bacteraemia. Int J Antimicrob Agents. 2020;55:105832.
Kishii K, Ito T, Watanabe S, Okuzumi K, Hiramatsu K. Recurrence of heterogeneous methicillin-resistant Staphylococcus aureus (MRSA) among the MRSA clinical isolates in a Japanese university hospital. J Antimicrob Chemother. 2008;62:324–8.
Ma W, Sun J, Yang S, Zhang L. Epidemiological and clinical features for cefepime heteroresistant Escherichia coli infections in Southwest China. Eur J Clin Microbiol Infect Dis. 2016;35:571–8.
Bryson D, Hettle AG, Boraston AB, Hobbs JK. Clinical mutations that partially activate the stringent response confer multidrug tolerance in Staphylococcus aureus. Antimicrob Agents Ch. 2020;64:e02103–19.
Hamad MA, Austin CR, Stewart AL, Higgins M, Vazquez-Torres A, Voskuil MI. Adaptation and Antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Ch. 2011;55:3313–23.
Hemsley CM, Luo JX, Andreae CA, Butler CS, Soyer OS, Titball RW. Bacterial drug tolerance under clinical conditions is governed by anaerobic adaptation but not anaerobic respiration. Antimicrob Agents Ch. 2014;58:5775–83.
Zhang SS, Liu S, Wu N, Yuan YH, Zhang WH, Zhang Y. Small non-coding RNA RyhB mediates persistence to multiple antibiotics and stresses in Uropathogenic Escherichia coli by reducing cellular metabolism. Front Microbiol. 2018;9:1–10.
Abraham EP. A glimpse of the early history of the cephalosporins. Rev Infect Dis. 1979;1:99–105.
Bo G. Giuseppe Brotzu and the discovery of cephalosporins. Clin Microbiol Infect. 2000;6:6–9.
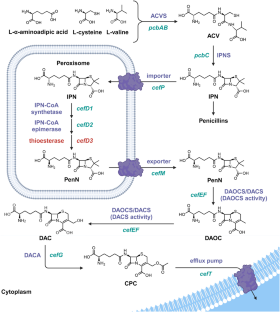
Brakhage AA. Molecular regulation of beta-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev. 1998;62:547–85.
Hu Y, Zhu B. Study on genetic engineering of Acremonium chrysogenum, the cephalosporin C producer. Synth Syst Biotechnol. 2016;1:143–9.
Abraham E. Selective reminiscences of beta-lactam antibiotics: early research on penicillin and cephalosporins. Bioessays. 1990;12:601–6.
Abraham EP, Newton GGF, Hale CW. Purification and some properties of Cephalosporin-N, a new Penicillin. Biochem J. 1954;58:94–102.
Hamilton-Miller JMT. Sir Edward Abraham's contribution to the development of the cephalosporins: a reassessment. Int J Antimicrob Ag. 2000;15:179–84.
Hamilton-Miller JMT. The cephalosporins and Sir Edward Abraham: Recollections about a great scientist and his part in the discovery of these antibiotics. J Antibiot. 2000;53:1003–7.
Hodgkin DC, Maslen EN. The x-ray analysis of the structure of cephalosporin C. Biochem J. 1961;79:393–402.
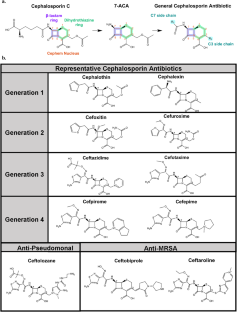
Elks J. Structural formulae and nomenclature of the cephalosporin antibiotics. Drugs. 1987;34:240–6.
Lin XM, Kuck U. Cephalosporins as key lead generation beta-lactam antibiotics. Appl Microbiol Biot. 2022;106:8007–20.
Byford MF, Baldwin JE, Shiau CY, Schofield CJ. The mechanism of ACV Synthetase. Chem Rev. 1997;97:2631–50.
Baldwin JE, Gagnon J, Ting HH. N-Terminal Amino-acid sequence and some properties of Isopenicillin-N Synthetase from Cephalosporium-Acremonium. Febs Lett. 1985;188:253–6.
Pang CP, et al. Purification of isopenicillin N synthetase. Biochem J. 1984;222:789–95.
Randall CR, et al. X-ray-absorption studies of the ferrous active-site of Isopenicillin N-Synthase and related model complexes. Biochem-Us. 1993;32:6664–73.
Roach PL, et al. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature. 1997;387:827–30.
Ullan RV, Casqueiro J, Banuelos O, Fernandez FJ, Gutierrez S, Martin JF. A novel epimerization system in fungal secondary metabolism involved in the conversion of isopenicillin N into penicillin N in Acremonium chrysogenum. J Biol Chem. 2002;277:46216–25.
Brewer SJ, Farthing JE, Turner MK. The oxygenation of the 3-methyl group of 7beta-(5-D-aminoadipamido)-3-methylceph-3-em-4-carboxylic acid (desacetoxycephalosporin C) by extracts of Acremonium chrysogenum [proceedings]. Biochem Soc Trans. 1977;5:1024–6.
Dotzlaf JE, Yeh WK. Copurification and characterization of deacetoxycephalosporin C synthetase/hydroxylase from Cephalosporium acremonium. J Bacteriol. 1987;169:1611–8.
Higgens CE, Hamill RL, Sands TH, Hoehn MM, Davis NE. Letter: The occurrence of deacetoxycephalosporin C in fungi and streptomycetes. J Antibiot. 1974;27:298–300.
Rabe P, Kamps J, Schofield CJ, Lohans CT. Roles of 2-oxoglutarate oxygenases and isopenicillin N synthase in beta-lactam biosynthesis. Nat Prod Rep. 2018;35:735–56.
Samson SM, et al. Cloning and expression of the fungal expandase hydroxylase gene involved in cephalosporin biosynthesis. Bio-Technol. 1987;5:1207. +
Kupka J, Shen YQ, Wolfe S, Demain AL. Studies on the ring-cyclization and ring-expansion enzymes of beta-lactam biosynthesis in Cephalosporium acremonium. Can J Microbiol. 1983;29:488–96.
Lejon S, Ellis J, Valegard K. The last step in cephalosporin C formation revealed: crystal structures of deacetylcephalosporin C acetyltransferase from Acremonium chrysogenum in complexes with reaction intermediates. J Mol Biol. 2008;377:935–44.
Schmitt EK, Hoff B, Kuck U. Regulation of cephalosporin biosynthesis. Adv Biochem Eng Biotechnol. 2004;88:1–43.
Gutierrez S, Diez B, Montenegro E, Martin JF. Characterization of the Cephalosporium acremonium pcbAB gene encoding alpha-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase: linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol. 1991;173:2354–65.
Martin JF, Ullan RV, Casqueiro J. Novel genes involved in cephalosporin biosynthesis: the three-component isopenicillin N epimerase system. Adv Biochem Eng Biotechnol. 2004;88:91–109.
Gutierrez S, Velasco J, Fernandez FJ, Martin JF. The Cefg gene of Cephalosporium-Acremonium is linked to the Cefef gene and encodes a Deacetylcephalosporin-C Acetyltransferase closely related to Homoserine O-Acetyltransferase. J Bacteriol. 1992;174:3056–64.
Teijeira F, Ullan RV, Guerra SM, Garcia-Estrada C, Vaca I, Martin JF. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem J. 2009;418:113–24.
Ullan RV, Teijeira F, Guerra SM, Vaca I, Martin JF. Characterization of a novel peroxisome membrane protein essential for conversion of isopenicillin N into cephalosporin C. Biochem J. 2010;432:227–36.
Ullan RV, Liu G, Casqueiro J, Gutierrez S, Banuelos O, Martin JF. The cefT gene of Acremonium chrysogenum C10 encodes a putative multidrug efflux pump protein that significantly increases cephalosporin C production. Mol Genet Genom. 2002;267:673–83.
Gutierrez S, Fierro F, Casqueiro J, Martin JF. Gene organization and plasticity of the beta-lactam genes in different filamentous fungi. Anton Leeuw Int J G 1999;75:81–94.
Liu L, Chen Z, Liu W, Ke X, Tian X, Chu J. Cephalosporin C biosynthesis and fermentation in Acremonium chrysogenum. Appl Microbiol Biotechnol. 2022;106:6413–26.
Skatrud PL, Queener SW. An electrophoretic molecular karyotype for an industrial strain of Cephalosporium acremonium. Gene. 1989;78:331–8.
Smith DJ, et al. Beta-Lactam antibiotic biosynthetic genes have been conserved in clusters in Prokaryotes and Eukaryotes. Embo J. 1990;9:741–7.
Pollegioni L, Rosini E, Molla G. Cephalosporin C acylase: dream and(/or) reality. Appl Microbiol Biotechnol. 2013;97:2341–55.
Bianchi D, Bortolo R, Golini P, Cesti P. Enzymatic transformation of cephalosporin C to 7-ACA by simultaneous action of immobilized d-amino acid oxidase and glutaryl-7-ACA acylase. Appl Biochem Biotech. 1998;73:257–68.
Elander RP. Industrial production of beta-lactam antibiotics. Appl Microbiol Biotechnol. 2003;61:385–92.
Bush K, Bradford PA. beta-Lactams and beta-Lactamase Inhibitors: An Overview. Cold Spring Harb Perspect Med. 2016;6:a025247.
Chaudhry SB, Veve MP, Wagner JL. Cephalosporins: A focus on side chains and beta-lactam cross-reactivity. Pharmacy. 2019;7:103.
Giamarellou H. Fourth generation cephalosporins in the antimicrobial chemotherapy of surgical infections. J Chemother. 1999;11:486–93.
Comments
Post a Comment